Metagenome sequencing technology has been widely used in the study of microorganisms. Researchers use computational methods to reconstruct a large number of microbial genomes from humans, animals and the environment from sequencing data. The common pipeline includes assembly and binning. The currently widely used metagenomic binning methods are all unsupervised (not dependent on the reference genome) methods, ignoring the information from the reference genomes. The biomedical AI team of the ISTBI of Fudan University proposed a semi-supervised metagenomic binning tool, termed SemiBin (https://github.com/BigDataBiology/SemiBin), which uses a deep siamese neural network to learn the information from the reference genomes. The results show that SemiBin outperforms existing state-of-the-art binning methods in several different habitats from GMGCv1 (Global Microbial Gene Catalog), which the team had published last year.
This research was published in Nature Communications on April 28, 2022 ( https://www.nature.com/articles/s41467-022-29843-y).
SemiBin defines contigs from different species and genus as cannot-link pairs, while exploiting the structure of the problem (an approach termed self-supervision) to generate must-link constraints.. The input must-link and cannot-link constraints are used to train the siamese neural network.
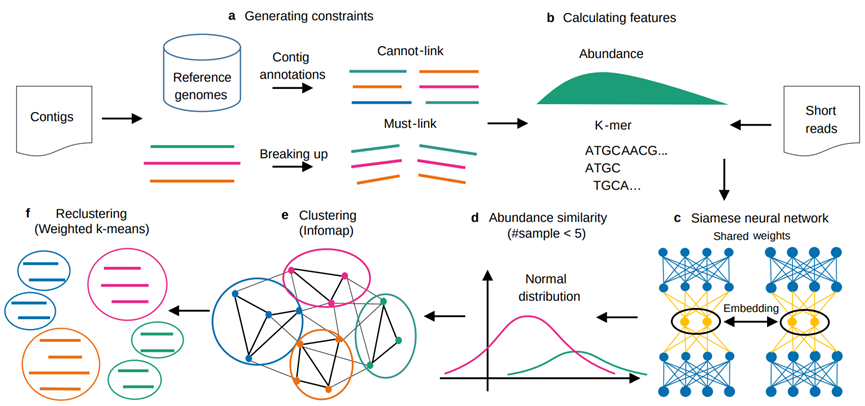
Fig.1 SemiBin pipeline
Researchers evaluated the results of SemiBin on several simulated and real datasets, including human gut, dog gut, ocean, and soil. In all cases, SemiBin could reconstruct more high-quality bins; with improvements ranging from 17.5% to 171.4%;see Figure 2).
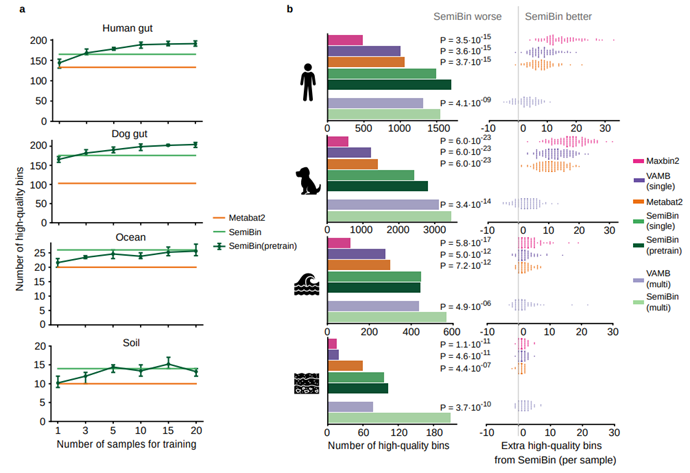
Fig.2 Benchmark results on real datasets
SemiBin has been able to obtain good binning results, but contig annotations and model training still require a lot of computing resources. Therefore, we proposed SemiBin (pretrain): pretraining model from multiple samples, and then directly applying the model to other samples. Compared with Metabat2, SemiBin (pretrain) can reconstruct 60.4%, 99.2%, 48.0% and 74.6% more high-quality bins. Based on GMGC, we proposed a total of 10 environment pre-training models, and these pre-training models are better than Metabat2 (Figure 3). The pre-trained model enables SemiBin to be directly used in large-scale data analysis, providing the possibility for a better understanding of microorganisms in the environment.
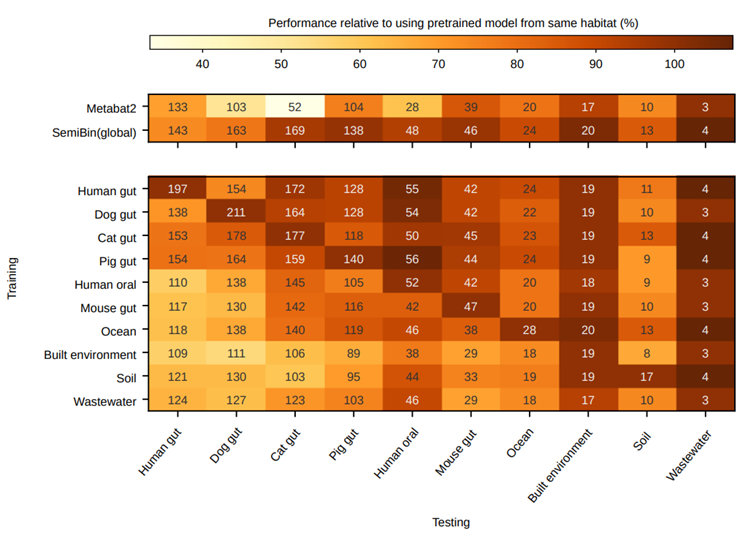
Fig. 3 Pretrained models from 10 environments
Since publication, work has continued and new releases have improved the algorithms to produce better results faster by exploiting self-supervised learning. A manuscript describing these improvements will be submitted in early 2023.
Shaojun Pan from the Biomedical AI Team of the ISTBI, Fudan University is the first author of this research, and Professor Xing-Ming Zhao and Luis Pedro Coelho are co-corresponding authors.

