Neurological and psychiatric disorders have complicated etiologies and are difficult to treat or manage. Many of these disorders have overlapped phenotypic profiles, but the underlying mechanisms cannot be fully explained through known disease genes and genomic variants. Drs Xing-Ming Zhao and Jingqi Chen from the Biomedical-AI group in ISTBI, Fudan University have investigated the tissue-specific contributions of noncoding genomic variants to the shared mechanisms of neuropsychiatric disorders. They have constructed a comprehensive tissue-specific disorder similarity network, and found that non-brain tissues have their unique, indispensable roles in these disorders. This work has been published in Molecular Psychiatry as of April, 4th, 2022 (full text link:https://rdcu.be/cKDPd).

Figure 1. This work has been online since April 4th, 2022 (https://www.nature.com/articles/s41380-022-01529-3).
The researchers first collected genome-wide associated loci from multiple large databases for 14 types of neuropsychiatric disorders, including 9 psychiatric disorders such as Schizophrenia, and 5 neurological disorders such as Alzheimer's disease. To understand the roles of the noncoding variants in these disorders, the researchers designed a research procedure based on distal regulation and data integration of multiple biological networks, as shown in Figure 2. They identified the tissue-specific regulatory target genes for the noncoding variants in five tissues (cerebral cortex, hippocampus, small intestine, lung, and liver), and interpreted the tissue-specific biological pathways of non-coding variants involved in the 14 disorders. They have found that neurological disorders and psychiatric disorders have different pathways affected by the disrupted noncoding regulations in brain and non-brain tissues: the affected pathways in the brain are mainly related to nervous system development, while the affected pathways in non-brain tissues are more likely to be related to synapses and neuroinflammation; neurological disorders are more likely to affect neuroinflammatory pathways than psychiatric disorders, in both brain and non-brain tissues.
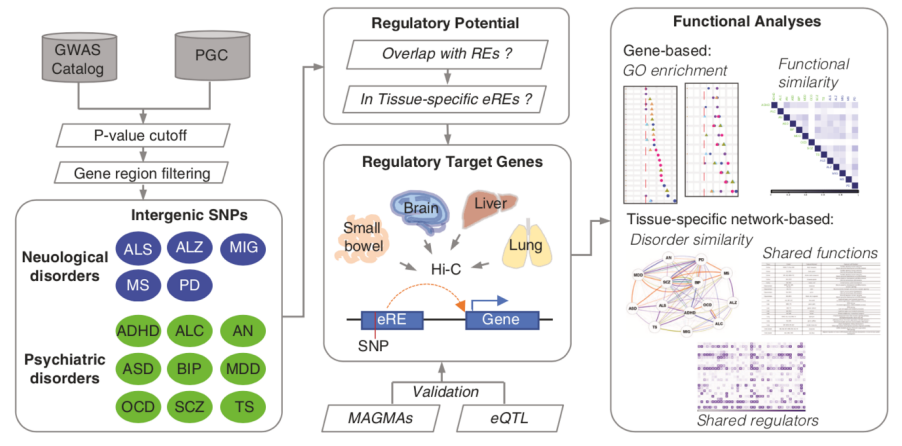
Figure 2. Mining of tissue-specific noncoding regulatory mechanisms for 14 neuropsychiatric disorders on 5 tissues. ALS: Amyotrophic lateral sclerosis; ALZ: Alzheimer's disease; MIG: Migraine; MS: Multiple sclerosis; PD: Parkinson's disease; ADHD: Attention deficit hyperactivity disorder; ALC: Alcohol dependence; AN: Anorexia nervosa; ASD: Autism spectrum disorder; BIP: Bipolar disorder; MDD: Major depressive disorder; OCD: Obsessive-compulsive disorder; SCZ: Schizophrenia; TS: Tourette syndrome.
Further, the researchers integrated tissue-specific regulatory target genes with gene co-expression networks of the five tissues to construct a disorder similarity network, as shown in Figure 3. Disorders that are conventionally known to be similar or related show strong similarities in a variety of tissues, such as Schizophrenia and Bipolar disorder in all five tissues. Neurodevelopmental disorders, such as Autism spectrum disorder, Attention deficit hyperactivity disorder, and Tourette syndrome, show strong similarities in brain tissues. There are also unexpected findings. For example, Parkinson's disease, a neurological disorder, show strong similarities with multiple psychiatric disorders in both brain and non-brain tissues. In fact, Parkinson's disease does have some clinical features of mental illness such as hallucinations. Parkinson's disease and Schizophrenia both affect the dopamine pathway, and Parkinson's disease has comorbidities with Schizophrenia and Major depressive disorder.
Based on the disorder similarity network in Figure 3, multiple disorders show strong similarities in the small intestine, in addition to in the brain tissues. Further integrating transcription factor regulatory networks, this study identified drug target transcription factors that may be shared by disorders in the same clusters, as shown in Figure 4. Some drug target transcription factors are specific to non-brain tissues, showing the potential importance of non-brain tissues in disorder management. For example, the target genes of the transcription factor NR1I3 are significantly enriched in the blue module of the small intestine, which corresponds to a cluster of disorders including Alcohol dependence, Migraine, and Obsessive-compulsive disorder, and this enrichment occurs only in the small intestine but not in the brain tissue. NR1I3 is the target of the approved drug atorvastatin known to treat Migraine, and the findings in this study suggest that the effects of this drug may be related to disease pathways in the small intestine, and that this drug may be tested for other disorders in the same cluster. The tissue-specific enrichment pattern of the drug target transcription factors may also suggest potential mechanisms of certain side effects. For instance, pseudoephedrine, commonly used to treat nasal and sinus congestion, has been reported to trigger bipolar manic episodes. Pseudoephedrine's target transcription factor is NFATC1, while the only module regulated by NFATC1 that contains Bipolar disorder is in the small intestine, suggesting that this side effect may be regulated through the small intestine.
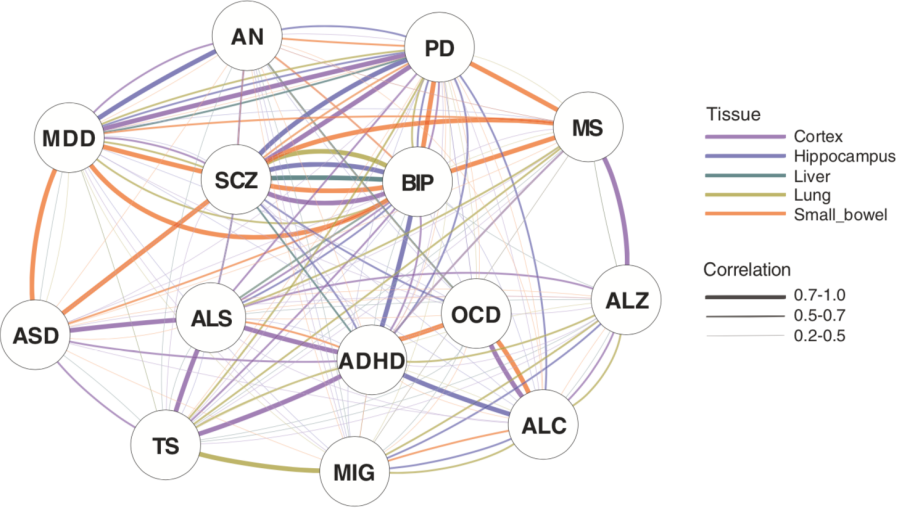
Figure 3. The tissue-specific disorder similarity network. Nodes represent disorders; different colors of edges represent different tissues; the thickness of edges represent the level of similarity (correlation).
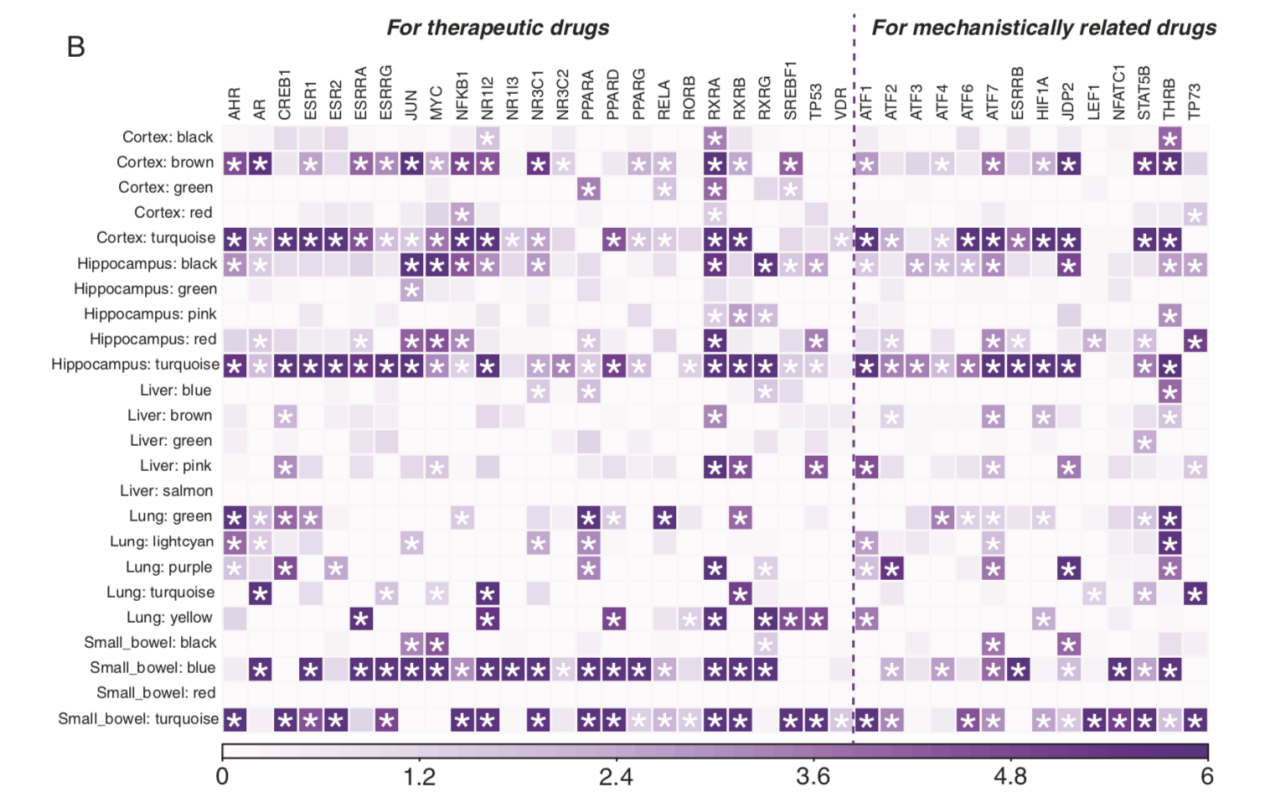
Figure 4. The enrichment patterns of drug target transcription factors in the modules of each tissue. Each row is a tissue-specific module, and each colume is a drug target transcription factor. The asterisk represents a significant enrichment of the target genes of a transcription factor in a module, and the shade of color represents the degree of enrichment (-log10 FDR).
This study innovatively explored the contributions of noncoding variants to the shared mechanisms of multiple neuropsychiatric disorders from the perspective of multi-tissue specificity, revealing the indispensable roles of non-brain tissues in the process of these disorders. This study may provide new ideas and potential drug targets for future development of drugs and systematic interventions for the neuropsychiatric disorders.
Dr. Jingqi Chen, a young associate researcher in the biomedical AI team of the Institute of Science and Technology for Brain-inspired Intelligence in Fudan University, is the first author of this study. Drs. Xing-Ming Zhao and Jingqi Chen are co-corresponding authors. This work is supported by the National Key R&D Program of China, the National Natural Science Foundation of China, the 111 Project of China, the Shanghai Municipal Science and Technology Major Project, Natural Science Foundation of Shanghai, ZJ Lab, and Shanghai Center for Brain Science and Brain-Inspired Technology.

