Recently, research teams lead by Prof. Shanfeng Zhu at ISTBI developed a deep learning-based model, DeepMHCII, for accurate MHC-II peptide binding affinity prediction, which can be further used for clinical TCR-based epitope identification. The study was published in Bioinformatics (ISMB2022) on 27 June 2022.
Major histocompatibility complex (MHC) molecules play an important role in T cell-mediated adaptive immune responses. MHC molecules first bind peptide fragments from pathogens and are then presented on the surface of antigen-presenting cells (APCs). Following recognition of the MHC-peptide complex by the T cell receptor (TCR), the adaptive immune response is activated to fight and eliminate the invading pathogen. Inspired by the biological background knowledge that the binding affinity of MHC-II molecular binding peptides is mainly determined by the binding core of peptides, DeepMHCII proposed a novel binding interaction convolution layer, which allows to integrate all potential binding cores with the MHC pseudo sequence, through modeling the interaction with multiple convolutional kernels. The convolutional layer that models the direct interactive information greatly improves the accuracy of model prediction.
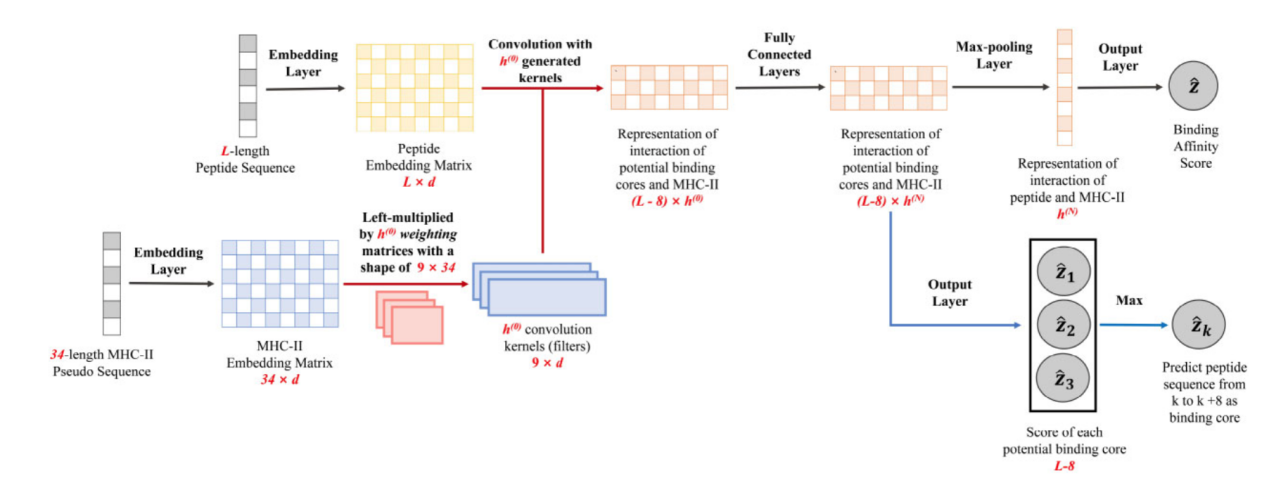
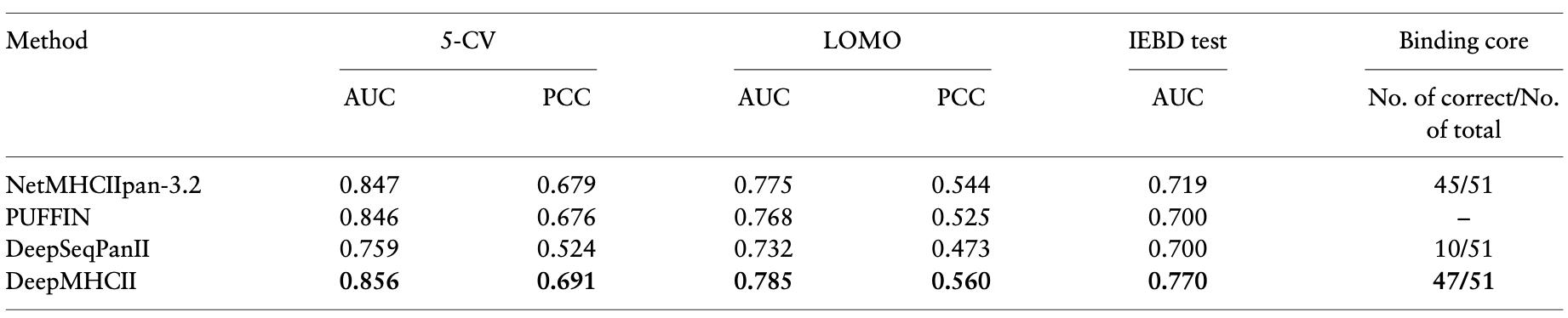
Extensive empirical experiments with four large-scale datasets demonstrate that DeepMHCII significantly outperformed four state-of-the-art methods under numerous settings, such as 5-fold cross-validation, leave one molecule out, validation with independent testing sets and binding core prediction. All these results and visualization of the predicted binding cores indicate the effectiveness of DeepMHCII, and the importance of properly modeling biological facts in deep learning for high predictive performance and efficient knowledge discovery.
DeepMHCII is publicly available at https://github.com/yourh/DeepMHCII.
Full Article:
You R, Qu W, Mamitsuka H, Zhu S(*). DeepMHCII: a novel binding core-aware deep interaction model for accurate MHC-II peptide binding affinity prediction. Bioinformatics (ISMB2022) 38 (Suppl 1):i220-i228. doi: 10.1093/bioinformatics/btac225.

