Multimorbidity refers to two or more diseases that co-occur in one patient not by chance. Compared with single disease, multimorbidity usually reduces the patients’ quality of life and increases their mortality and economic expenditures. Multimorbidity is usually caused by the interactions between environment and genetics, and its mechanism is very complex. In the past ten years, genome-wide association studies (GWAS) have found that there are shared genetic risks among a few multimorbid disease-pairs. However, due to the lack of large-scale diagnosis and genetic data of the same cohort, the genetic pattern among multimorbidities remain unclear. Over the past decade, the UK Biobank has collected electronic health records and genomic data of about 500,000 people, making it possible to integrate diagnostic data and genomic data of the same samples to explore the genetic pattern of multimorbidities among common diseases.
Recently, the bioinformatics group of Drs. Xing-Ming Zhao and Jingqi Chen from the Institute of Science and Technology for Brain-Inspired Intelligence (ISTBI) of Fudan University, analyzed the shared genetic risks of multimorbidities among common diseases. They found that the hub diseases and molecules in the genetically interpretable multimorbidity network may be the key factors for the treatment of multimorbidity. On July 5, 2021, this study was published online in the Journal ofGenome Medicinewith the title of "A global overview of genetically interpretable diseases among common diseases in the UK Biobank".
Based on the electronic health record data of the UK Biobank, the study found 11,285 multimorbid disease-pairs among 438 common diseases. In addition to the prevelant multimorbidities affecting the same physiological system, the study found additional 41 physiological category-pairs of different physiological systems where the multimorbidity relationships were also more prevelant than expected. Metabolic diseases have the highest multimorbidity rate of cross-physiological systems. Besides, most of the cross-category multimorbidities are mediated by a few diseases. For example, the multimorbidities between nutritional and psychiatric, joint and spine categories are mainly mediated by the obesity.
Based on the GWAS summary statistics of the UK Biobank, the researchers found that nearly half (47%) of the multimorbidities can be explained by genetic factors at either the loci, network or overall genetic structure level. The genetically interpretable multimorbidities show different genetic patterns in terms of the physiological systems: the multimorbidities affecting the same physiological system tend to be explained by the loci-level genetic components, while the multimorbidities affecting different physiological systems tend to be explained by the network-level genetic components. This result suggests that multimorbidities affecting the same and different physiological systems may have different biological origins - the former tends to be directly derived from pleiotropic loci, and the latter tends to be indirectly derived from the same or similar biological functions.
Through the functional analysis of the molecules and networks shared by multimorbidities, researchers found that a small number of genetic components can explain a large portion of multimorbidities, and these common genetic components are mainly enriched in functions of cellular immunity, protein metabolism and gene silencing. The researchers further constructed two multimorbidity networks by linking multimorbid disease-pairs interpretable through the loci- and network-level genetic components. Through complex network analysis, they found that there were obvious module structures in the two multimorbidity networks. Besides, they identified the hub diseases and molecules for each multimorbidity module. These hub diseases and molecules can help understand the etiology of multimorbidity. For example, tobacco dependence, five respiratory diseases (emphysema, empyema, respiratory failure, chronic obstructive pulmonary disease, pneumonia), three cardiovascular diseases (atherosclerosis, aortic aneurysm and dissection, peripheral vascular disease), and lung cancer, form a multimorbidity module, among which tobacco dependence is the hub disease. The most common genes in this module areIREB2andCHRNA3, in whichIREB2encodes an iron response element binding protein (IRP) that regulates iron metabolism, and CHRNA3 encodes neuronal nicotinic acetylcholine receptor. The results suggest that iron metabolism and neuronal nicotinic acetylcholine receptor may be the main targets to study the multimorbidity of tobacco dependence with these diseases.
This study firstly integrates the electronic health records and genomic data of the same samples to systematically analyze the genetic patterns of multimorbidities among common diseases. This study explored the trend and genetic pattern of multimorbidity of same and different physiological systems, and pointed out that hub diseases and the shared biomolecules may be the key to the treatment of multimorbidity. An online database has been extablished to facilitate researchers and doctors to browse, search or download these multimorbidities (https://multimorbidity.comp-sysbio.org).
Guiying Dong, a PhD student from the ISTBI of Fudan University, is the first author of this paper. Professor Xing-Ming Zhao and young associate researcher Jingqi Chen are the co-corresponding authors. The research was supported by the Shanghai Municipal Science Technology Major Project, National Key R&D Program of China, National Natural Science Foundation of China, the 111 Project of China, Natural Science Foundation of Shanghai, ZJ Lab, and Shanghai Center for Brain Science and Brain-Inspired Technology.
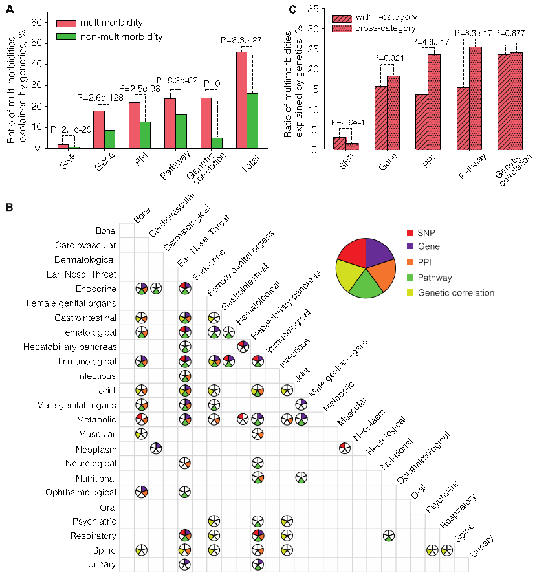
Fig 1. Genetically interpretable multimorbidities.
A.The proportion of multimorbidities and non-multimorbidity explained by single nucleotide polymorphisms (SNPs), gene,s protein-protein interactions (PPIs), biological pathways and genetic correlations.
B.Multimorbidity of different categories that are significantly explained by genetic components. Each circle is divided into five parts, representing five types of genetic components. The color filled parts of each circle indicate that this type of genetic components can significantly explain the multimorbidity of the category.
C.The proportion of multimorbidities of same and different physiological systems that can be explained by single nucleotide polymorphisms (SNPs), genes, protein-protein interactions (PPIs), pathways and genetic correlations.
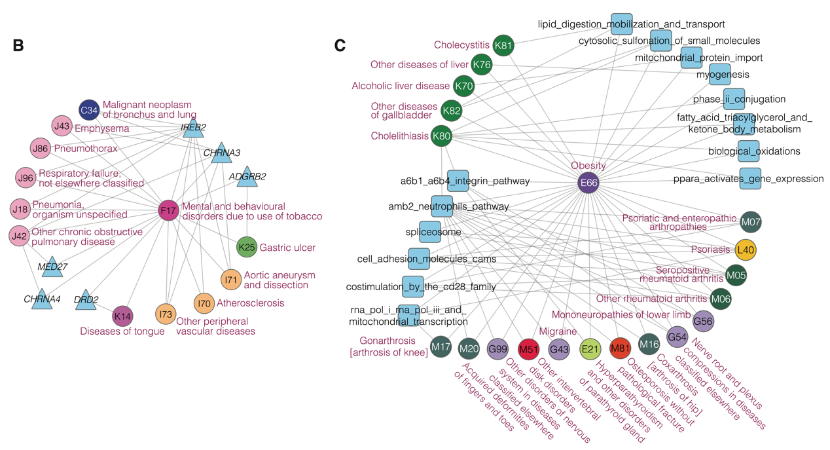
Fig 2. Case study of genetically interpretable multimorbidity networks. Circles, triangles and squares represent disease, gene and pathways, respectively. The color of the circle (disease) corresponds to the category of the disease, and the color code is the same as that in Fig 1.
B.Multimorbidities and shared genes related to hub disease F17-tobacco dependence in the LG-Module 5.
C.Multimorbidities and shared pathways related to hub disease E66-Obesity in the NG-Module 1 and 6. Only the top five pathways are shown.

