Cell fate is jointly determined by the intracellular molecular regulatory network and the extracellular microenvironment. Single-cell omics technology can measure molecular expression profiles in cells, but loses spatial location information. In recent years, the rapid development of spatial omics technologies has enabled researchers to obtain the spatial information in addition to measuring rich molecular expression profiles.
Mainstream spatial omics technologies include spatial transcriptomics, spatial proteomics, spatial metabolomics, etc. With the development of these technologies, a large amount of spatial omics data is rapidly generated and maintained in various heterogeneous data platforms. Before specific data analysis, researchers must first download the raw data and then process it correctly into a standard format, which is usually time-consuming and laborious. Another problem is the amount of data, because some advanced experimental techniques currently provide large fields of view and high spatial resolution (such as Stereo-seq, MERFISH, etc.), processing these data requires a lot of memory and time consumption.
The latest research of Zhiyuan Yuan from the ISTBI of Fudan University, Professor Michael Q. Zhang from the University of Texas at Dallas, and Dr. Jianhua Yao from Tencent AI Lab have given a solution to support users to quickly browse, visualize, and load the spatial data of interest. The relevant research were recently published in Nature Methods entitled "SODB facilitates comprehensive exploration of spatial omics data".
SODB provides data from 26 spatial omics technologies, with a data volume of more than 50 million cells (spots). All data are processed into Anndata format by standard procedures, and are compatible with various analysis software such as SCANPY and Squidpy. SODB also provides a variety of data analysis and visualization modules, including gene spatial expression, cell type annotation, gene expression comparison, SOView visualization analysis, etc. (Figure 2). In addition, the supporting Python toolkit pysodb is also provided, which can easily read data with only one line of code. Compared with traditional data processing and loading methods, it brings improved time efficiency and memory efficiency. Taking Slide-seq data as an example, the time efficiency is increased by 160 times (the traditional method takes 19.04 minutes to read, and pysodb only needs 7.16 seconds); the memory efficiency is increased by 549 times (the traditional method requires 21.97GB, and pysodb only needs 0.04GB peak memory).
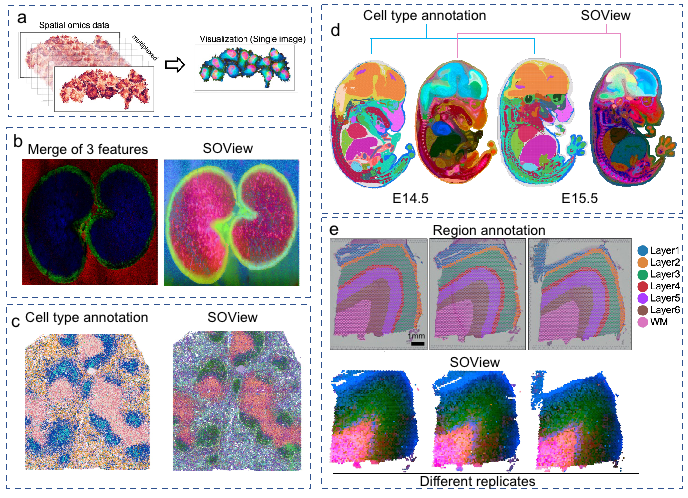
Figure 2. SOView visualization module
Currently, SODB is open to everyone, and the data can be accessed through the website [https://gene.ai.tencent.com/SpatialOmics/] or the Python package [https://github.com/TencentAILabHealthcare/pysodb].
The co-corresponding authors of this project are Zhiyuan Yuan (and the first author) of ISTBI of Fudan University, Dr. Jianhua Yao of Tencent AI Lab, and Professor Michael Q. Zhang of the University of Texas at Dallas. The co-first author is Wentao Pan of Tsinghua University. Professor YiZhao from the Institute of Computing Technology, Chinese Academy of Sciences, Professor Xiu Li from Tsinghua University, Xuan Zhao and Zhimeng Xu of Tencent AI Lab, and other collaborators have made significant contributions to this project.
This project was supported by the Shanghai Municipal Scienceand Technology Major Project (no. 2018SHZDZX01), ZJ Laboratory, Shanghai Center for Brain Science and Brain-Inspired Technology and 111 Project (no. B18015).
Zhiyuan Yuan, received his Ph.D. from Tsinghua University in June 2022, under the supervision of Professor Michael Q. Zhang. In September 2022, he joined the ISTBI, Fudan University as a young associate researcher. Mainly engaged in bioinformatics, especially the research and application of spatial omics computing methods, mainly involving theories including deep learning, statistical modeling and probabilistic graphical models, and application scenarios include the construction of large-scale brain spatiotemporal maps, brain diseases and tumor microenvironment space-time modeling. In recent years, he has published several papers in Nature Methods (2021, 2023), Nature Communications (2022a, 2022b), Nucleic Acids Research (2022).

